Work Package 5 - AeroDAT
In WP5 (AeroDAT), headed by HMGU, Aerosol and biological response DATa obtained in WP2–WP4 will be integrated and jointly analyzed in order to identify the aerosol parameters that are critical for the health effects. The overall aim is to improve risk management by identifying the components that likely contribute the most to development of adverse health outcomes. This WP will heavily use the bioinformatics services at the Weizmann Institute. This will provide identifying guidelines for which sources to regulate in order to optimize public health benefits. The toxicological results will be integrated with the ‘omics and epigenetic data, while the biological responses will be put in context to the exposure data (i.e. the comprehensive analysis of the chemical and physical properties of the exposure aerosol) to link aerosol sources via observed biological effects in the cell cultures with potential heath-effects in humans. All partners will contribute to WP 5. For this decisive task, bioinformatics and chemometric expertise will be bundled and accompanied by software developing capacity. With the ever increasing complexity and volume of data in life sciences in general, and in genomics in particular, it is clear that analysis methods constantly need to be developed and adapted. The ICB pioneers in this challenging field and contributes constantly to the scientific community. We are part of various consortia and for instance recently launched and coordinate a single genomics network in Germany. Furthermore, we coordinate the project 'Sparse2big' involving eight Helmholtz Centers with the goal to develop, evaluate and share methods for data imputation and integration.
RESEARCH METHODOLOGY AND APPROACH OF AeroDAT
The Objectives of WP5 are the establishment of an interpretable integrative network linking toxicity parameter with molecular omics data and delineating toxicity effects of cell culture and 3D models on pathway level. WP5 supports individual WPs for the analysis of all aeroHEALTH generated omics dat with state-of-the-art processing, quality assessment and normalization.
To develop a method to integrate toxicity with molecular measurements we use regression analysis and mixed effect modeling allowing accounting for technical covariates. We use existing data and synthetic data to develop and fine-tune the methods, in order to apply the methods to aeroHEALTH data once it becomes available. For in the integration epigenetic, transcriptomic and proteomic data, we use prior knowledge to allocate interaction pairs subjected to correlation analysis. The multi-omics network will enrich the aeroHEALTH knowledge graph of toxicity effects. Integrated data analysis will identify the biological effects in multi-omics data through a combination of similarity networks based on omics-data profiles, independent component analysis (ICA) and machine learning (ML), allowing the deconvolution of mixed signals from the multi-cell cultures. The extracted features will then be used as inputs to ML techniques, e.g., random forests, for predicting biological effects of air pollution. Methods bridging the "omics" findings to specific mechanism of toxicity will be applied to strengthen link between particle composition/source apportionment and human health effects. By this, further steps towards a cell-culture result based risk analysis will be done. During the course of the project, in-depth investigations of biological associations between the newly obtained cell culture-based results and animal model data are performed.
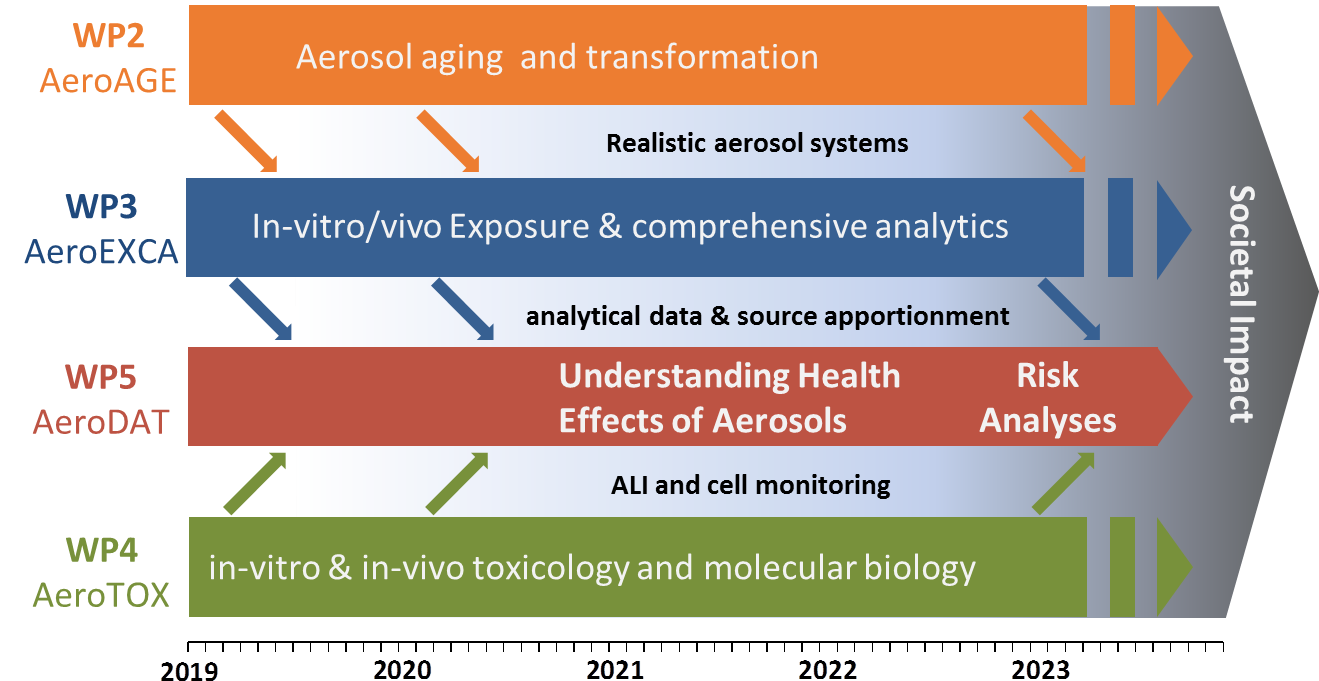
The general interaction structure of the work packages as well as the flow of information is depicted in the figure below. The WP5 is central in the way that data from all WP are jointly analyzed here. All partners are contributing to WP5 using the mathematical expertise and "big-data"-infrastructure at HMGU (ICB).